Research
The life of a cell emerges in the forms of the various biomolecular self-assemblies on precise interactions at all levels of complexities, spanning different time and length scales. Cellular information, for example, is transferred through the mechanism by which proteins, nucleic acids, metabolites, and other biomolecules bind and/or assemble into active complexes to initiate specific biological tasks (e.g., gene transcription and regulation, signaling, immune and stress responses). Although the structure, dynamics, and function of these processes can be studied through a variety of biophysical, biochemical, and structural biology techniques, models and/or theories constructed, based on the information from experiments, are still far from being complete (not see the wood for the trees). Computer simulation offers a unique means for exact results for the models we build, so as to bridge the connection between experiment and theory. Currently, there are several challenges.
Protein-DNA Interactions
protein-DNA interactions
Molecular mechanism of gene regulation.
“Life is nothing but a competition to be the criminal rather than the victim.”
Try to manipulate it ! (move, rotate etc.) Below is a NGL app showing Fis molecule (PDBID: 3IV5), which is a nucleoid-associated protein in E-Coli. It regulates the expression of gene through binding DNA.
“For competition, we must substitute cooperation. We must learn to think of the human race as one family.”
Tsai et al., J. Am. Chem. Soc. 2019
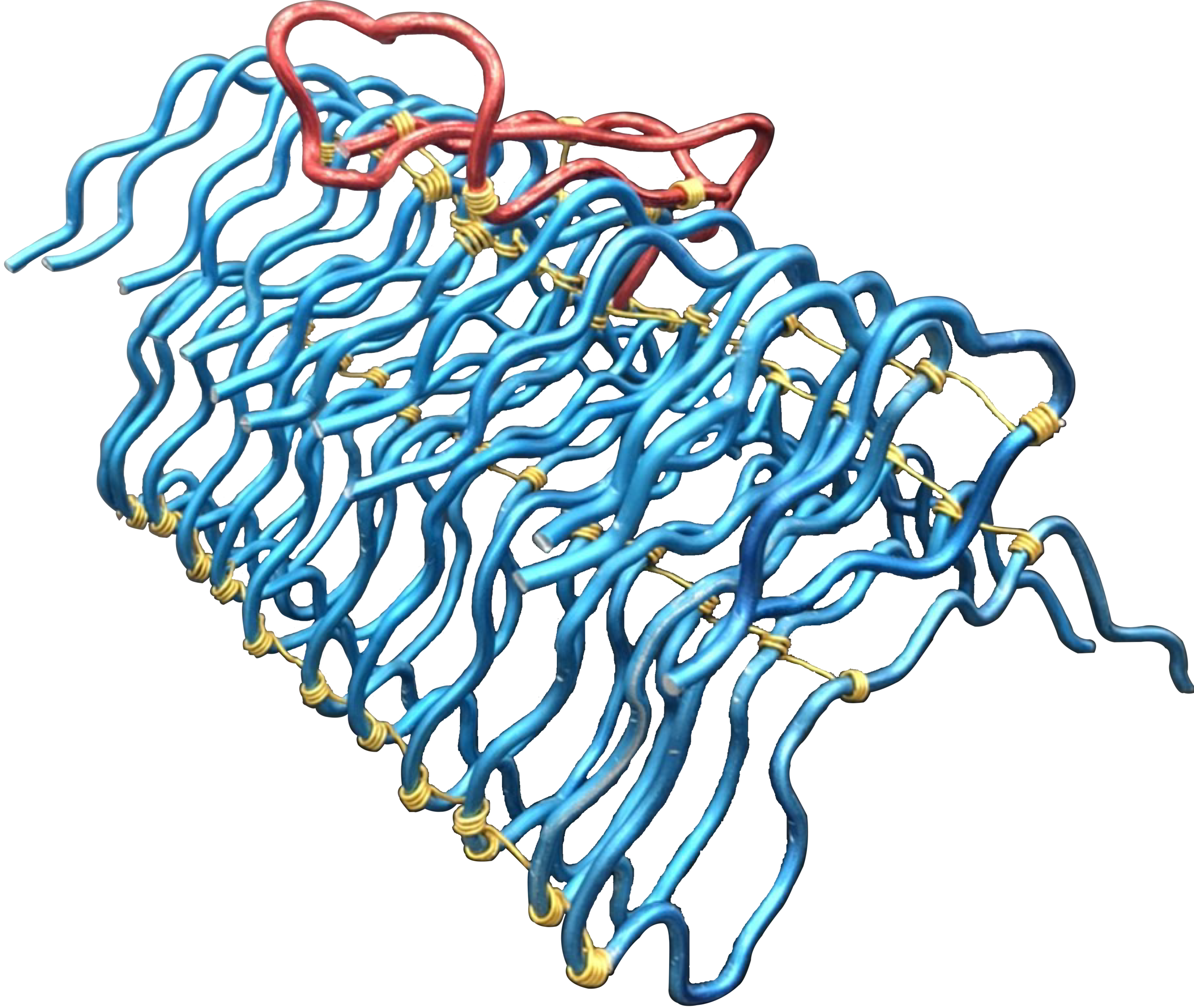
Fibril Surface-templated Aggregation in vivo
Stochastic fluctuation
plays an important role in aggregation kinetics. The reaction mechanism correlates with the magnitude of the stochastic fluctuations.
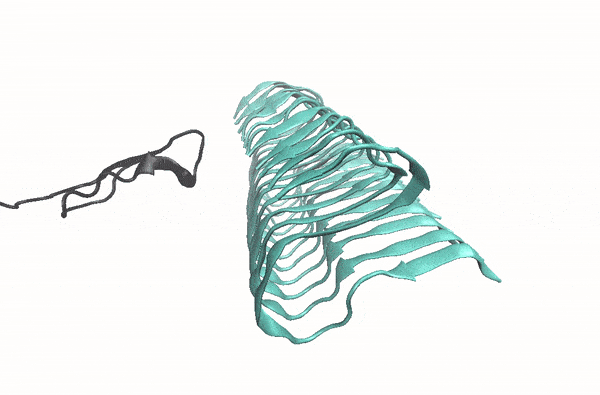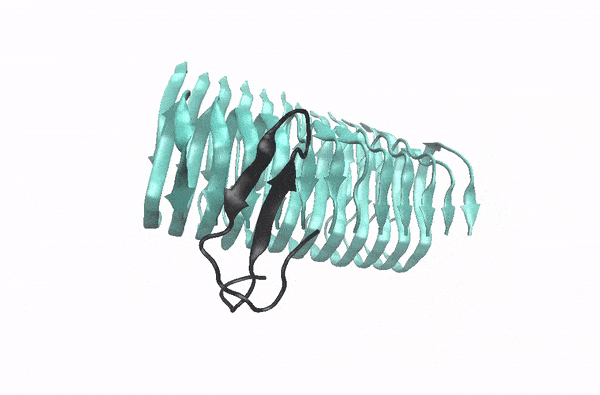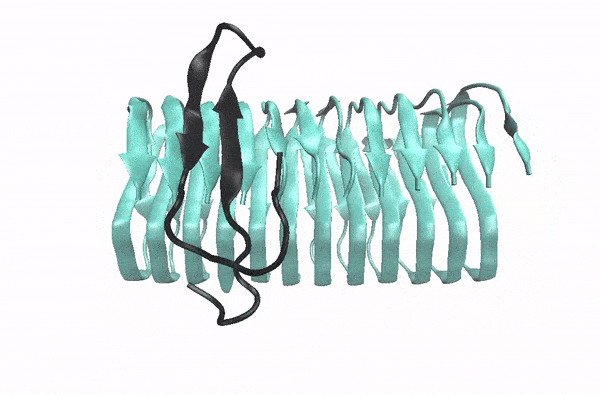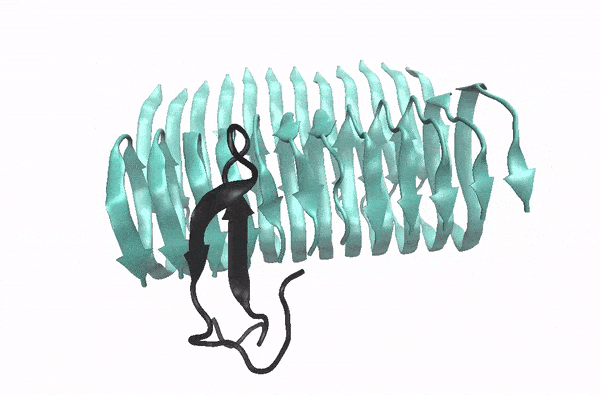
Abeta monomer searching over the surface of a fibril
This movie illustrates how an Abeta monomer (black) uses its hydrophobic patch to identify the C-terminal surface of a fibril (cyan)—it forms a “ C-terminal amyloid precursor”.
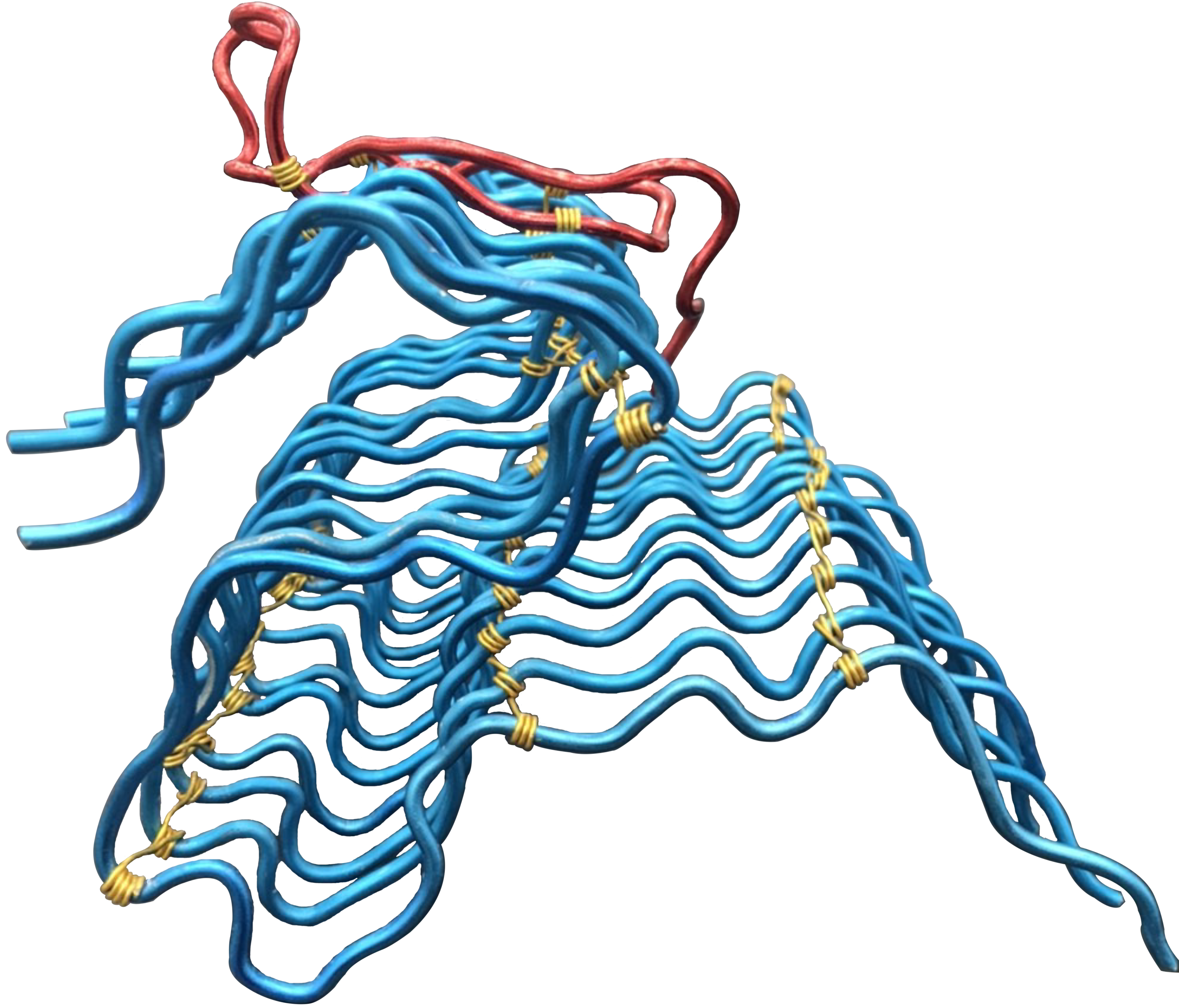
Fibrillar twisting
is an emergent mechanical property arising from a collective motion of connected neighboring chains. (Please wait for about 20s to see the changes)
Tsai et al., Protein Sci. 2016
protein functional binding and assemble
Post-translational modification and Role of electrostatics in protein-protein interaction.
Protein folding
Multi-scale folding-unfolding dynamics of globular proteins & peptides.