Selected Publications (as pi)
*Corresponding author
†Contributed equally to this work
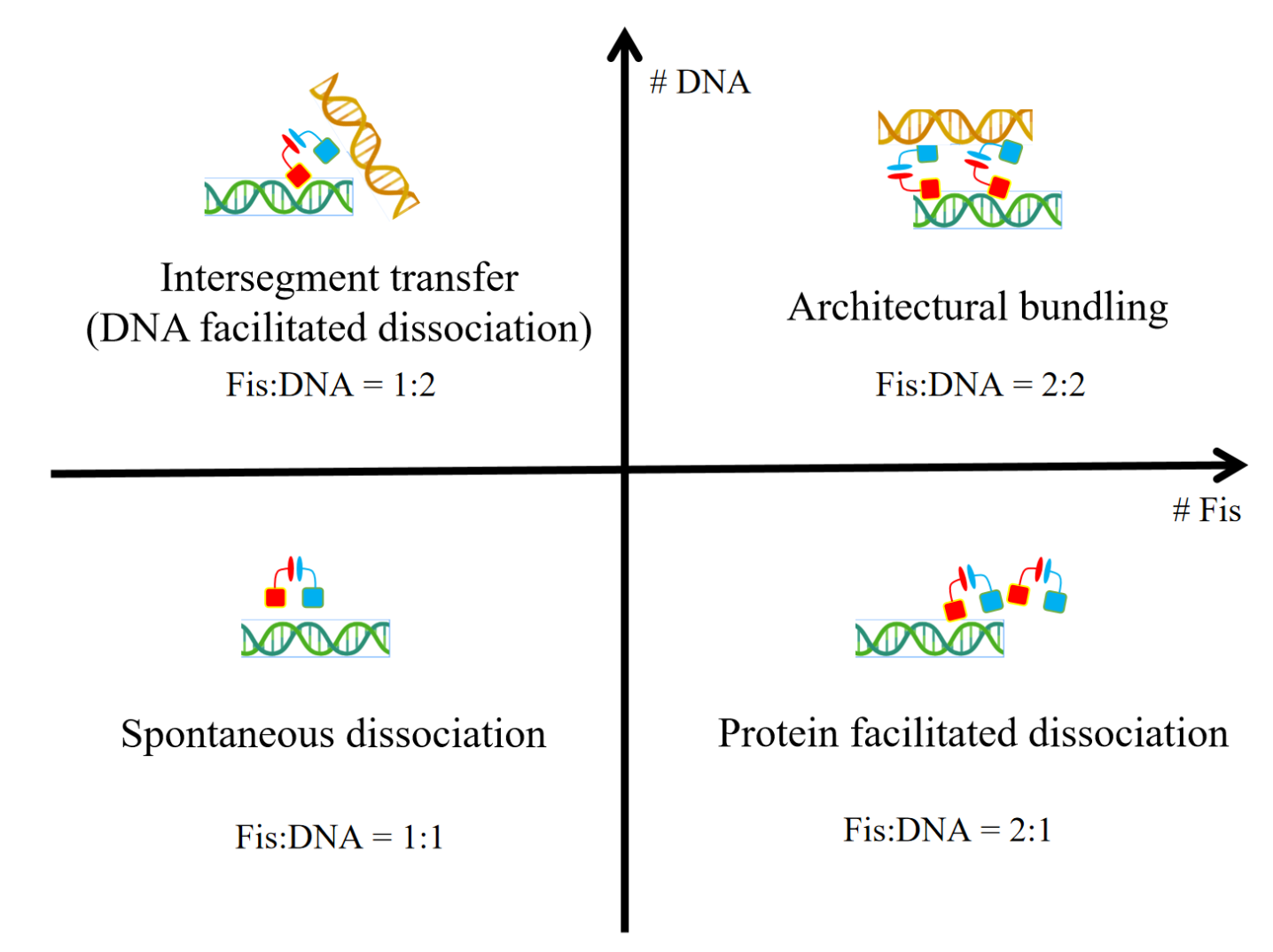
Chen, X.*; Jin, S.; Liu, C.-H.; Levy, Y.*; Tsai, M.-Y.*; Wolynes, P. G.* Intersegment Transfer and the Dynamical Architecture of Fis Protein-DNA Multimer Complexes. J. Am. Chem. Soc. 2025, 147 (33), 30277–30286. (https://doi.org/10.1021/jacs.5c09262)
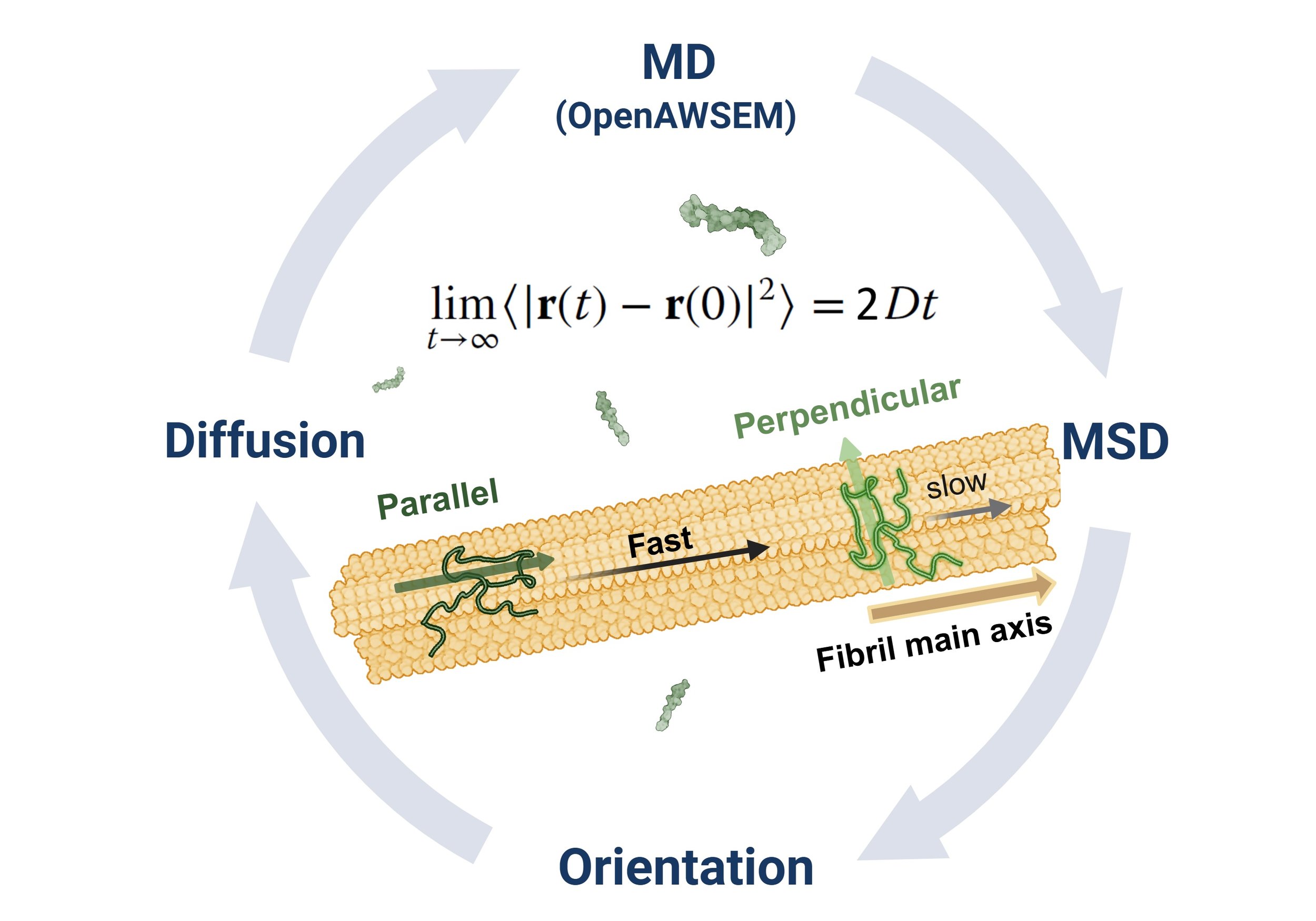
Ma, Y.-W.†; Wang, G.-F.†; Chen, H.-Y.; Tsai, M.-Y.* Exploring Abeta42 Monomer Diffusion Dynamics on Fibril Surfaces through Molecular Simulations. Protein Sci. 2025, 34 (6), e70131. (https://onlinelibrary.wiley.com/doi/full/10.1002/pro.70131)
Li, Chengxuan; Wei, Tingyi; Cheung, Margaret S; Tsai, M.-Y.* Deciphering the Cofilin Oligomers via Intermolecular Disulfide Bond Formation: A Coarse-grained Molecular Dynamics Approach to Understanding Cofilin’s Regulation on Actin Filaments. J. Phys. Chem. B 2024, 128 (19), 4590–4601. (https://pubs.acs.org/doi/abs/10.1021/acs.jpcb.3c07938)
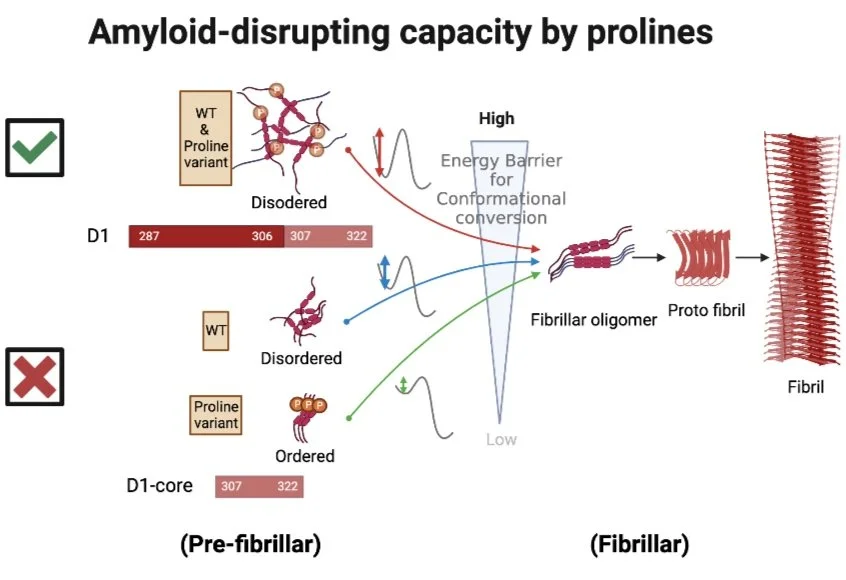
Lin, T.-Y.; Ma, Y.-W.; Tsai, M.-Y.* Early-stage Oligomerization of Prion-like Polypeptides Reveals the Molecular Mechanism of Amyloid-disrupting Capacity by Proline Residues. J. Phys. Chem. B. 2023, 127 (5), 1074-1088. (https://pubs.acs.org/doi/10.1021/acs.jpcb.2c05463)
Chen, X.; Tsai, M.-Y.*; Wolynes, P. G.* The Role of Charge Density Coupled DNA Bending in Transcription Factor Sequence Binding Specificity: A Generic Mechanism for Indirect Readout. J. Am. Chem. Soc. 2022, 144 (4), 1835–1845. (https://pubs.acs.org/doi/10.1021/jacs.1c11911)
Ma, Y.-W.; Lin, T.-Y.; Tsai, M.-Y.* Fibril Surface-dependent Amyloid Precursors revealed by Coarse-grained Molecular Dynamics Simulation. Front. Mol. Biosci. 2021, 8, 719320.
(https://doi.org/10.3389/fmolb.2021.719320)Tsai, M.-Y.*,†;Shen, J.-L.†; Schafer, N. P.; Wolynes, P. G.*, Modeling Protein Aggregation Kinetics: The Method of Second Stochasticization. J. Phys. Chem. B 2021, 125 (4), 1118–1133. (https://pubs.acs.org/doi/10.1021/acs.jpcb.0c10331)
Full Publications List
Journal Articles (Sorted by Year)
CCU (2023.2 - Present)
Submitted
Ma, Yuan-Wei; Wang, Guan-Fang; Tsai, M.-Y.* Exploring Abeta42 Monomer Diffusive Dynamics on Fibril Surfaces through Molecular Simulations. (BioRxiv)
Manuscripts in preparation
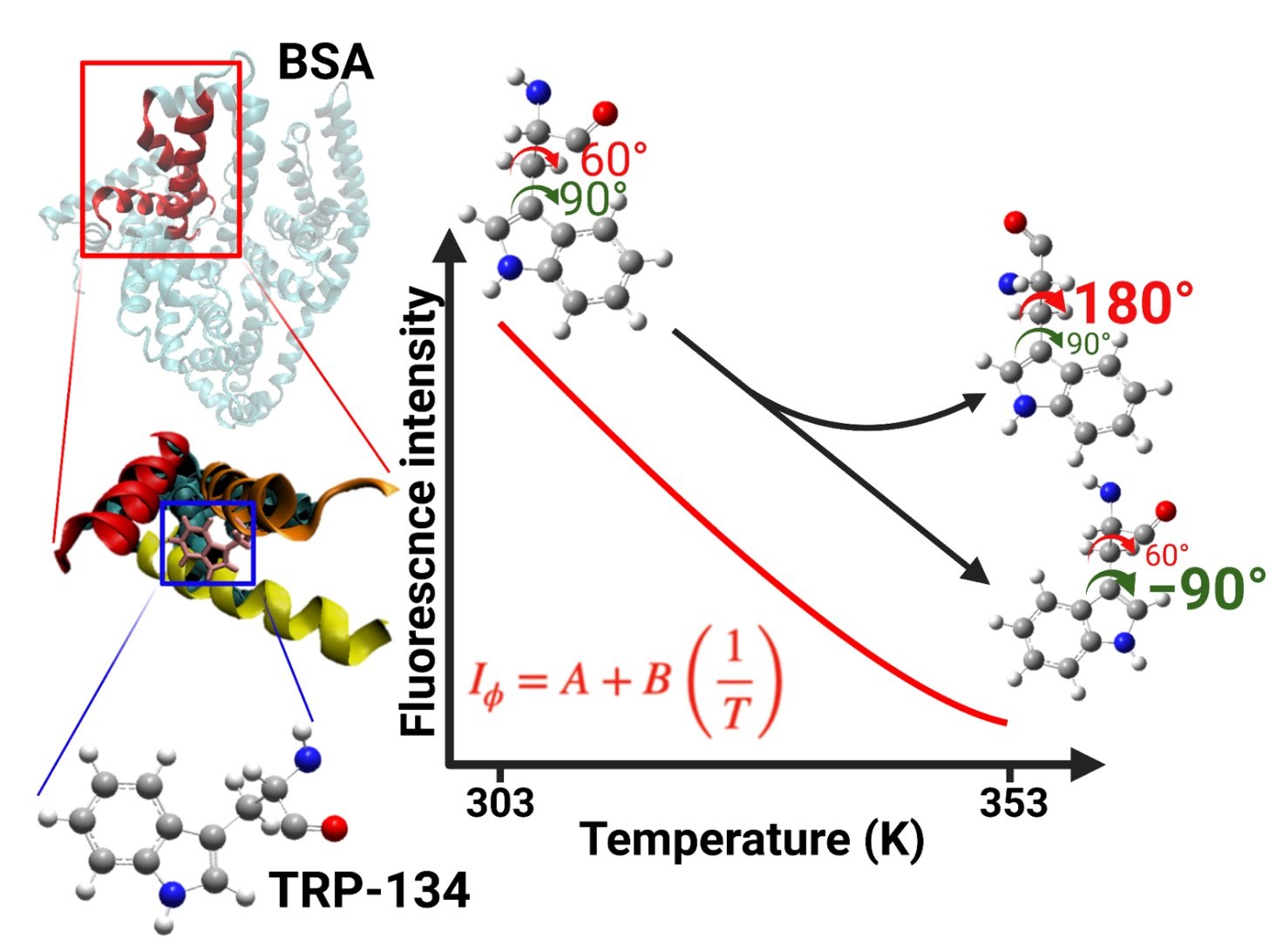
Chou, Yun-Chu; Yung-Ting Lee; Lin, Tong-You; Tsai, M.-Y.* Unraveling the Intricacies of Tryptophan's Fluorescence in Proteins: Exploring Tryptophan Rotamers through Atomistic Simulations.
Published
1. Chen, X.*; Jin, S.; Liu, C.-H.; Levy, Y.*; Tsai, M.-Y.*; Wolynes, P. G.* Intersegment Transfer and the Dynamical Architecture of Fis Protein-DNA Multimer Complexes. J. Am. Chem. Soc. 2025, 147 (33), 30277–30286. (https://doi.org/10.1021/jacs.5c09262)
2. Ma, Y.-W.†; Wang, G.-F.†; Chen, H.-Y.; Tsai, M.-Y.* Exploring Abeta42 Monomer Diffusion Dynamics on Fibril Surfaces through Molecular Simulations. Protein Sci. 2025, 34 (6), e70131. (https://onlinelibrary.wiley.com/doi/full/10.1002/pro.70131)
3. Tsai, M.-Y.* How simple models explain complex protein folding behaviors. J. Chin, Chem. Soc. 2025 Early view (https://doi.org/10.1002/jccs.202400396)
4. Li, Chengxuan; Wei, Tingyi; Cheung, Margaret S; Tsai, M.-Y.* Deciphering the Cofilin Oligomers via Intermolecular Disulfide Bond Formation: A Coarse-grained Molecular Dynamics Approach to Understanding Cofilin’s Regulation on Actin Filaments. J. Phys. Chem. B 2024, 128 (19), 4590–4601. (https://pubs.acs.org/doi/abs/10.1021/acs.jpcb.3c07938)
5. Chou, T.-C.; Hu, Y.-L.; Xie, G.-C.; Jiang, J.-C.; Peng, L.-Y.; Tsai, H.-C.; Yao, C.-T.; Tsai, Y.-J.; Huang, T.-Y.; Hu, J.-W.; Chen, Y.-C.; Tsai, M.-Y.; Chen, Y.-W.; Pan, P.-S. The Use of Multicomponent Reactions in the Development of Bis-Boronic Acids for the Detection of β-Sialic Acid. Org. Biomol. Chem. 2024, 22 (8), 1639–1645. (https://pubs.rsc.org/en/content/articlelanding/2024/ob/d3ob01877f)
TKU (2018.8 - 2023.1)
1. Lin, T.-Y.; Ma, Y.-W.; Tsai, M.-Y.* Early-stage Oligomerization of Prion-like Polypeptides Reveals the Molecular Mechanism of Amyloid-disrupting Capacity by Proline Residues. J. Phys. Chem. B. 2023, 127 (5), 1074-1088. (https://pubs.acs.org/doi/10.1021/acs.jpcb.2c05463)
2. Chou, Yun-Chu; Lin, Tong-You; Tsai, M.-Y.* A Rotamer Model to Explain the Temperature Dependence of BSA Protein Fluorescence. J. Chem. Chem. Soc. 2023, 70 (3), 386-393. (https://onlinelibrary.wiley.com/doi/abs/10.1002/jccs.202200528)
3. Chen, X.; Tsai, M.-Y.*; Wolynes, P. G.* The Role of Charge Density Coupled DNA Bending in Transcription Factor Sequence Binding Specificity: A Generic Mechanism for Indirect Readout. J. Am. Chem. Soc. 2022, 144 (4), 1835–1845. (https://pubs.acs.org/doi/10.1021/jacs.1c11911)
4. Chen, X.; Lu, W.; Tsai, M.-Y.; Jin, S.; Wolynes, P. G. Exploring the Folding Energy Landscapes of Heme Proteins Using a Hybrid AWSEM-Heme Model. J. Biol. Phys. 2022. (https://doi.org/10.1007/s10867-021-09596-3)
5. Ma, Y.-W.; Lin, T.-Y.; Tsai, M.-Y.* Fibril Surface-dependent Amyloid Precursors revealed by Coarse-grained Molecular Dynamics Simulation. Front. Mol. Biosci. 2021, 8, 719320. (https://doi.org/10.3389/fmolb.2021.719320)
6. Tsai, M.-Y.*,†;Shen, J.-L.†; Schafer, N. P.; Wolynes, P. G.*, Modeling Protein Aggregation Kinetics: The Method of Second Stochasticization. J. Phys. Chem. B 2021, 125 (4), 1118–1133. (https://pubs.acs.org/doi/10.1021/acs.jpcb.0c10331)
7. Jin, S.; Contessoto, V. S.; Chen, M.; Schafer, N. P.; Lu, W.; Chen, X.; Bueno, C.; Hajitaheri, A.; Sirovetz, B. J.; Davtyan, A.; Papoian, G. A.; Tsai, M.-Y.; Wolynes, P. G.* AWSEM-Suite: A Protein Structure Prediction Server Based On Template-guided, Coevolutionary-enhanced Optimized Folding Landscapes. Nucleic Acids Res. 2020, 48 (W1), W25-W30.(https://doi.org/10.1093/nar/gkaa356)
8. Tsai, M.-Y.; Zheng, W.; Chen, M.; Wolynes, P. G.* Multiple Binding Configurations of Fis Protein Pairs on DNA: Facilitated Dissociation versus Cooperative Dissociation. J. Am. Chem. Soc. 2019, 141 (45), 18113−18126. (https://pubs.acs.org/doi/10.1021/jacs.9b08287)
9. Tsai, M.-Y.* Role of Physical Nucleation Theory in Understanding Conformational Conversion between Pathogenic and Nonpathogenic Aggregates of Low-Complexity Amyloid Peptides. Res. Chem. Intermed. 2019, 45, 5357-5373. (https://link.springer.com/article/10.1007/s11164-019-03974-2)
Before TKU (before 2018.8)
Lin, X.; Kulkarni, P.; Bocci, F.; Schafer, N. P.; Roy, S.; Tsai, M.-Y.; He, Y.; Chen, Y.; Rajagopalan, K.; Mooney, S. M.; et al. Structural and Dynamical Order of a Disordered Protein: Molecular Insights into Conformational Switching of PAGE4 at the Systems Level. Biomolecules 2019, 9 (2), 77.
Lin, X.; Roy, S.; Jolly, M. K.; Bocci, F.; Schafer, N. P.; Tsai, M.-Y.; Chen, Y.; He, Y.; Grishaev, A.; Weninger, K.; et al. PAGE4 and Conformational Switching: Insights from Molecular Dynamics Simulations and Implications for Prostate Cancer. J. Mol. Biol. 2018, 430 (16), 2422–2438.
Zheng, W.; Tsai, M.-Y.; Wolynes, P. G.* Comparing the Aggregation Free Energy Landscapes of Amyloid Beta(1-42) and Amyloid Beta(1-40). J. Am. Chem. Soc. 2017, 139 (46), 16666–16676.
Chen, M.; Tsai, M.-Y.; Zheng, W.; Wolynes, P. G.* The Aggregation Free Energy Landscapes of Polyglutamine Repeats. J. Am. Chem. Soc. 2016, 138 (46), 15197–15203.
Zheng, W.; Tsai, M.-Y.; Chen, M.; Wolynes, P. G.* Exploring the Aggregation Free Energy Landscape of the Amyloid-β Protein (1-40). Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (42), 11835–11840.
Tsai, M.-Y.; Zhang, B.; Zheng, W.; Wolynes, P. G.* Molecular Mechanism of Facilitated Dissociation of Fis Protein from DNA. J. Am. Chem. Soc. 2016, 138 (41), 13497−13500.
Parra, R. G.; Schafer, N. P.; Radusky, L. G.; Tsai, M.-Y.; Guzovsky, A. B.; Wolynes, P. G.; Ferreiro, D. U.* Protein Frustratometer 2: A Tool to Localize Energetic Frustration in Protein Molecules, Now with Electrostatics. Nucleic Acids Res. 2016, 44 (W1), W356–W360.
Shiu, Y.-J.; Hayashi, M.; Shih, O.; Su, C.; Tsai, M.-Y.; Yeh, Y.-Q.; Su, C.-J.; Huang, Y.-S.; Lin, S.-H.; Jeng, U.-S.* Intrinsic Coordination for Revealing Local Structural Changes in Protein Folding-Unfolding. Phys. Chem. Chem. Phys. 2016, 18 (4), 3179–3187.
Tsai, M.-Y.; Zheng, W.; Balamurugan, D.; Schafer, N. P.; Kim, B. L.; Cheung, M. S.; Wolynes, P. G.* Electrostatics, Structure Prediction, and the Energy Landscapes for Protein Folding and Binding. Protein Sci. 2016, 25 (1), 255–269.
Tsai, M.-Y.*; Yuan, J.-M.; Lin, S.-H. Thermodynamic Insight into Protein Aggregation Using a Kinetic Ising Model. J Chin Chem Soc 2015, 62 (1), 21–25.
Tsai, M.-Y.*; Yuan, J.-M.; Yamaki, M.; Lin, C.-K.; Lin, S. H. Molecular Dynamics Insight into the Diverse Thermodynamic Behavior of a Beta-Hairpin Peptide. J Chin Chem Soc 2013, 60 (7), 915–928.
Lin, C.-K.*; Shih, C.-C.; Niu, Y.; Tsai, M.-Y.; Shiu, Y.-J.; Zhu, C.; Hayashi, M.; Lin, S. H. Theoretical Study on Structure and Sum-Frequency Generation (SFG) Spectroscopy of Styrene–Graphene Adsorption System. J. Phys. Chem. C 2013, 117 (4), 1754–1760.
Tsai, M.-Y.*; Yuan, J.-M.; Teranishi, Y.; Lin, S. H. Thermodynamics of Protein Folding Using a Modified Wako-Saitô-Muñoz-Eaton Model. J. Biol. Phys. 2012, 38 (4), 543–571.
Tsai, M. Y.*; Morozov, A. N.; Chu, K. Y.; Lin, S. H. Molecular Dynamics Insight into the Role of Tertiary (foldon) Interactions on Unfolding in Cytochrome c. Chem. Phys. Lett. 2009, 475 (1), 111–115.
Morozov, A. N.*; Shiu, Y. J.; Liang, C. T.; Tsai, M. Y.; Lin, S. H. Nonadditive Interactions in Protein Folding: The Zipper Model of Cytochrome C. J. Biol. Phys. 2007, 33 (4), 255–270.
Book Chapters
Tsai, M.-Y.*; Yuan, J.-M.; Lin, S. H. Chapter 6: Thermodynamics and Kinetics of Protein Folding and Aggregation. In Biophysics and Biochemistry of Protein Aggregation: Experimental and Theoretical Studies on Folding, Misfolding, and Self-Assembly of Amyloidogenic Peptides; Yuan, J.-M., Zhou, H.-X., Eds.; World Scientific, 2017.